Xiao Chen is a Professor of the Laboratory of Marine Protozoan Biodiversity & Evolution, affiliated with Marine College at Shandong University. His group aims to delineate mechanisms underlying chromatin regulation and genome evolution using eukaryotic cell models.
Dr. Chen has been focusing on the epigenetic regulation mechanism of cellular homeostasis maintenance using eukaryotic models including ciliate. The findings are highlighted as follows:
1) Clarified the gene transcription pattern and omics basis for maintaining cellular homeostasis under environmental stress, laying the foundation for further exploration of its epigenetic regulation mechanism;
2) Elucidated the molecular mechanisms by which epigenetic factors alter gene transcription levels and regulate cellular homeostasis, expanding our understanding of the involvement of epigenetic factors in regulating cellular homeostasis;
3) Revealed the pathogenic mechanism of cellular homeostasis imbalance caused by chromatin dysregulation and confirmed the potential diagnostic and therapeutic value of epigenetic regulation of cellular homeostasis.
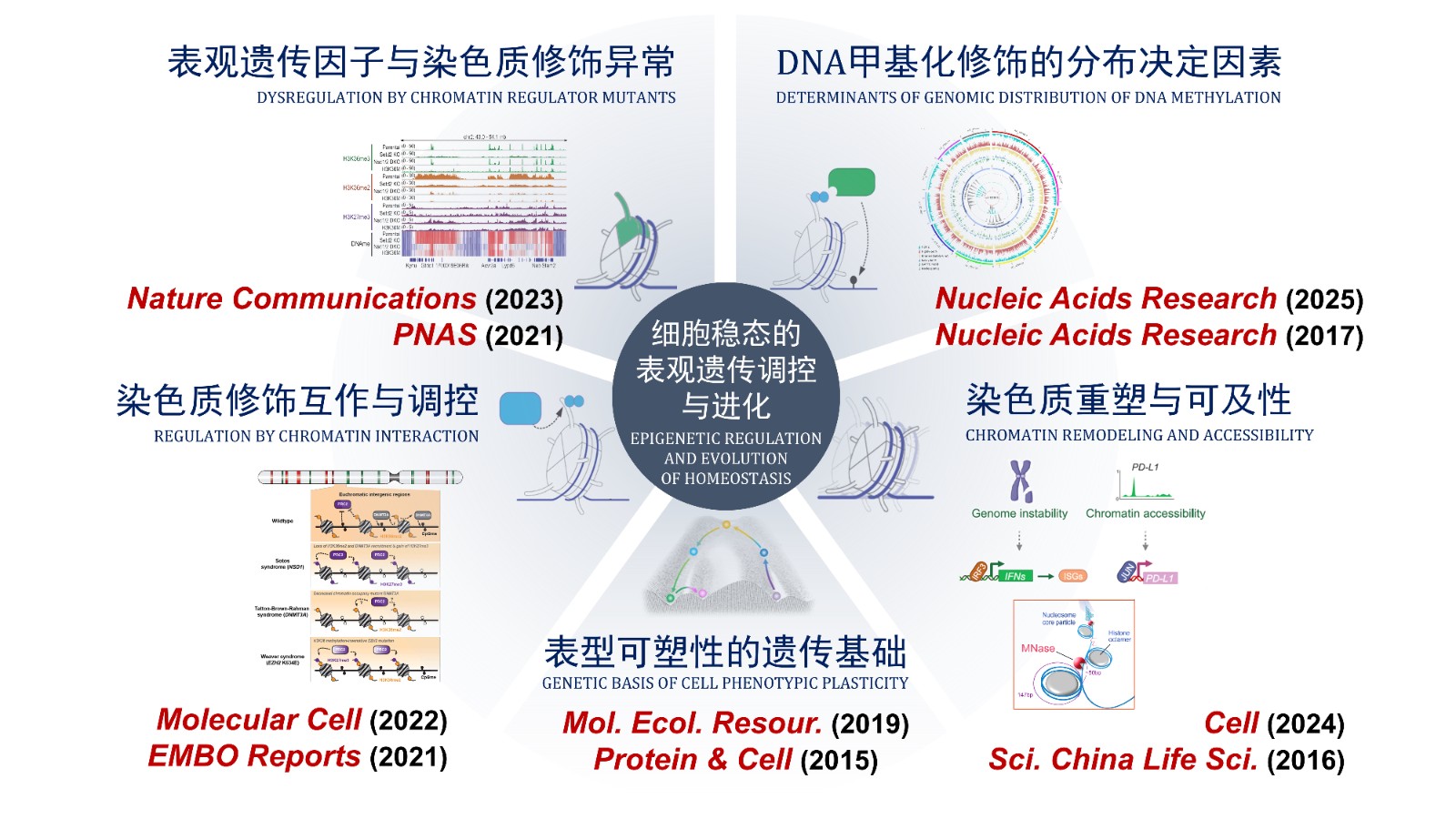
These findings have led to more than 40 publications in high-impact journals including Nature Communications, PNAS, and Molecular Cell. Dr. Chen's work was highly recognized by distinguished experts in chromatin fields as "a crucial discovery that will provide key information for current and future drug development" and "an important study that further illuminates on the role oncohistones have in gene transcription regulation".
Currently, the research on the dysregulation of gene transcription by chromatin states has been hampered by a lack of rationalized systems to dig deep into the underlying mechanism. Dr. Chen hereby aims to further explore the roles of histone modifications in regulating gene transcription and maintaining cell physiological and structural stability, in response to the key question of how unicellular eukaryotes maintain cellular homeostasis.
Profile: https://faculty.sdu.edu.cn/xc/en
Chen Lab: https://team.sdu.edu.cn/chenlab/en
Epigenetic Regulation and Evolution of Cellular Homeostasis
Multiple cancers and genetic disorders arise from the failure of cellular homeostasis mechanisms due to mutations in epigenetic factors. The greatest obstacle to treating these diseases currently lies in insufficient understanding of their homeostasis maintenance mechanisms. Enhancing our knowledge of the epigenetic regulatory mechanisms governing cellular homeostasis and their evolutionary trajectories will facilitate the discovery of novel therapeutic targets for related diseases, laying the foundation for developing efficient and reliable diagnostic and therapeutic strategies.
(1) [Nature, 2025] NSD2 targeting reverses plasticity and drug resistance in prostate cancer
(19) [PNAS, 2022] CHAF1A/B mediate silencing of unintegrated HIV-1 DNAs early in infection
(22) [Cell, 2021] Functional interrogation of DNA damage response variants with base editing screens
(31) [Cell Reports, 2020] The INO80 complex regulates epigenetic inheritance of heterochromatin
(46) [Protein & Cell, 2015] Phylogenomics of non-model ciliates based on transcriptomic analyses
2. 2024/01 - 2028/12 Program of Distinguished Young Scholar by Shandong University
3. 2023/01 - 2026/12 National Natural Science Foundation of China
4. 2023/01 - 2025/12 Youth Innovation Team of Shandong Provincial Higher Education Institutions
5. 2022/07 - 2025/06 Natural Science Foundation of Jiangsu Province
6. 2022/01 - 2027/01 Program of Qilu Young Scholar of Shandong University
1. 2023 National Outstanding Young Scientists (Overseas)
2. 2023 Distinguished Young Scholar by Shandong University
3. 2022 Qingdao Science and Technology Award (first prize of Natural Science Award)
4. 2021 Qilu Young Scholar by Shandong University
5. 2019 Outstanding Doctoral Dissertation by Shandong Province, China
6. 2017 Holz-Conner Award by International Society of Protistologists